Practice: Chp. 12#
source("iplot.R")
suppressPackageStartupMessages(library(rethinking))
12E1. What is the difference between an ordered categorical variable and an unordered one? Define and then give an example of each.
Answer. An ordered categorical variable is one in which the categories are ranked, but the distance between the categories is not necessarily constant. An example of an ordered categorical variable is the Likert scale; an example of an unordered categorical variable is the Blood type of a person.
See also Categorical variable and Ordinal data.
12E2. What kind of link function does an ordered logistic regression employ? How does it differ from an ordinary logit link?
Answer. It employs a cumulative link, which maps the probability of a category or any category of lower rank (the cumulative probability) to the log-cumulative-odds scale. The major difference from the ordinary logit link is the type of probabilties being mapped; the cumulative link maps cumulative probabilities to log-cumulative-odds and the ordinary logit link maps absolute probabilities to log-odds.
12E3. When count data are zero-inflated, using a model that ignores zero-inflation will tend to induce which kind of inferential error?
Answer. A ‘zero’ means that nothing happened; nothing can happen either because the rate of events is low because the process that generates events failed to get started. We may incorrectly infer the rate of events is lower than it is in reality, when in fact it never got started.
12E4. Over-dispersion is common in count data. Give an example of a natural process that might produce over-dispersed counts. Can you also give an example of a process that might produce under-dispersed counts?
Answer. The sex ratios of families actually skew toward either all boys or girls; see Overdispersion.
In contexts where the waiting time for a service is bounded (e.g. a store is only open a short time) but the model still uses a Poisson distribution you may see underdispersed counts because large values are simply not possible. If one queue feeds into another, and the first queue has a limited number of workers, the waiting times for the second queue may be underdispersed because of the homogeneity enforced by the bottleneck in the first queue. “)
display_markdown(” 12M1. At a certain university, employees are annually rated from 1 to 4 on their productivity, with 1 being least productive and 4 most productive. In a certain department at this certain university in a certain year, the numbers of employees receiving each rating were (from 1 to 4): 12, 36, 7, 41. Compute the log cumulative odds of each rating.
Answer. As a list:
freq_prod <- c(12, 36, 7, 41)
pr_k <- freq_prod / sum(freq_prod)
cum_pr_k <- cumsum(pr_k)
lco = list(log_cum_odds = logit(cum_pr_k))
display(lco)
- -1.94591014905531
- 0
- 0.293761118528163
- Inf
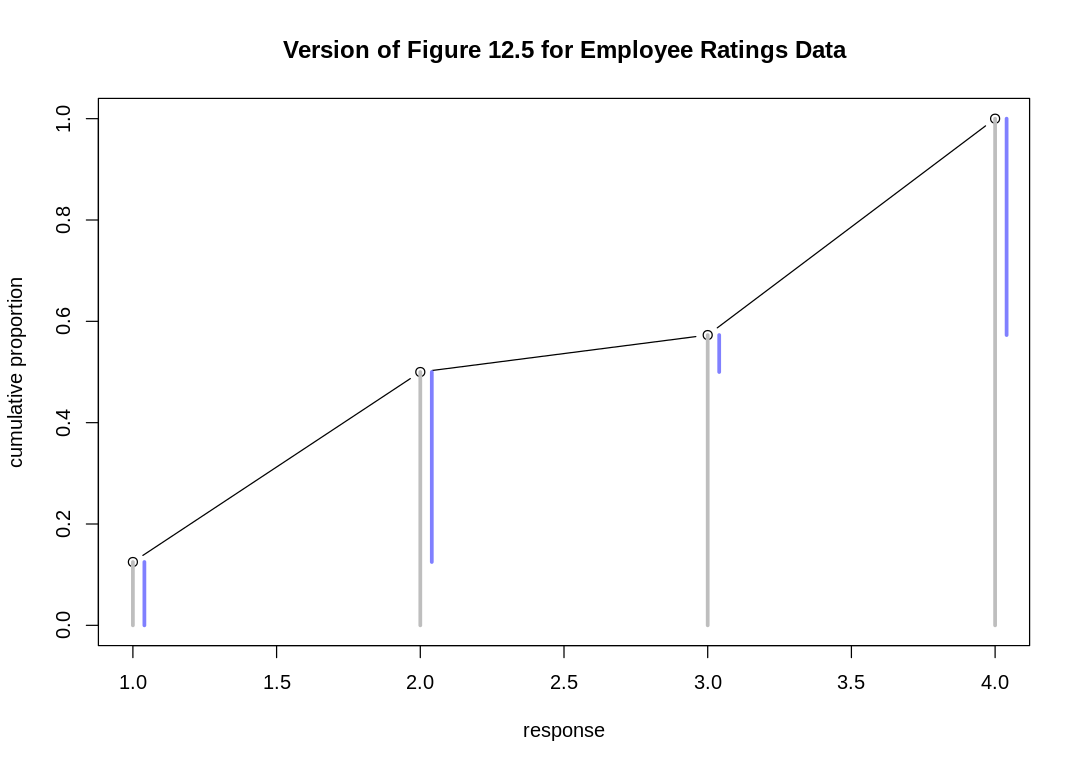
12M2. Make a version of Figure 12.5 for the employee ratings data given just above.
iplot(function() {
off <- 0.04
plot(
1:4, cum_pr_k, type="b",
main="Version of Figure 12.5 for Employee Ratings Data", xlab="response", ylab="cumulative proportion",
ylim=c(0,1)
)
# See `simplehist`
for (j in 1:4) lines(c(j,j), c(0,cum_pr_k[j]), lwd=3, col="gray")
for (j in 1:4) lines(c(j,j)+off, c(if (j==1) 0 else cum_pr_k[j-1],cum_pr_k[j]), lwd=3, col=rangi2)
})
12M3. Can you modify the derivation of the zero-inflated Poisson distribution (ZIPoisson) from the chapter to construct a zero-inflated binomial distribution?
Answer. Call \(p_z\) the additional probability of a zero, and \(p_b\) the probability of success associated with the binomial distribution. The likelihood of observing a zero is:
The likelihood of observing a non-zero value \(k\) is: $\( \begin{align} Pr(k | k>0,p_z,n,p_b) & = (1-p_z)Pr(k|n,p_b) \\ & = (1-p_z)\binom{n}{k}p_b^k(1-p_b)^{n-k} \end{align} \)$
To construct a GLM, use a logit link with \(p_z\) and \(p_b\) (in the typical case where \(n\) is known).
12H1. In 2014, a paper was published that was entitled “Female hurricanes are deadlier than male hurricanes.” As the title suggests, the paper claimed that hurricanes with female names have caused greater loss of life, and the explanation given is that people unconsciously rate female hurricanes as less dangerous and so are less likely to evacuate.
Statisticians severely criticized the paper after publication. Here, you’ll explore the complete data used in the paper and consider the hypothesis that hurricanes with female names are deadlier. Load the data with:
R code 12.38
library(rethinking)
data(Hurricanes)
Acquaint yourself with the columns by inspecting the help ?Hurricanes. In this problem, you’ll
focus on predicting deaths using femininity of each hurricane’s name. Fit and interpret the
simplest possible model, a Poisson model of deaths using femininity as a predictor. You can use
map or ulam. Compare the model to an intercept-only Poisson model of deaths. How strong is the
association between femininity of name and deaths? Which storms does the model fit (retrodict) well?
Which storms does it fit poorly?
Answer. The Hurricanes data.frame with an index and a standardized femininity:
data(Hurricanes)
hur <- Hurricanes
hur$index = 1:92
hur$StdFem = standardize(hur$femininity)
display(hur)
hur_dat = list(
Deaths = hur$deaths,
Femininity = hur$StdFem
)
m_hur_pois <- map(
alist(
Deaths ~ dpois(lambda),
log(lambda) <- a + bFemininity * Femininity,
a ~ dnorm(3, 0.5),
bFemininity ~ dnorm(0, 1.0)
),
data = hur_dat
)
m_hur_intercept <- map(
alist(
Deaths ~ dpois(lambda),
log(lambda) <- a,
a ~ dnorm(3, 0.5)
),
data = hur_dat
)
| name | year | deaths | category | min_pressure | damage_norm | female | femininity | index | StdFem |
|---|---|---|---|---|---|---|---|---|---|
| <fct> | <int> | <int> | <int> | <int> | <int> | <int> | <dbl> | <int> | <dbl> |
| Easy | 1950 | 2 | 3 | 960 | 1590 | 1 | 6.77778 | 1 | -0.0009347453 |
| King | 1950 | 4 | 3 | 955 | 5350 | 0 | 1.38889 | 2 | -1.6707580424 |
| Able | 1952 | 3 | 1 | 985 | 150 | 0 | 3.83333 | 3 | -0.9133139565 |
| Barbara | 1953 | 1 | 1 | 987 | 58 | 1 | 9.83333 | 4 | 0.9458703621 |
| Florence | 1953 | 0 | 1 | 985 | 15 | 1 | 8.33333 | 5 | 0.4810742824 |
| Carol | 1954 | 60 | 3 | 960 | 19321 | 1 | 8.11111 | 6 | 0.4122162926 |
| Edna | 1954 | 20 | 3 | 954 | 3230 | 1 | 8.55556 | 7 | 0.5499353710 |
| Hazel | 1954 | 20 | 4 | 938 | 24260 | 1 | 9.44444 | 8 | 0.8253673305 |
| Connie | 1955 | 0 | 3 | 962 | 2030 | 1 | 8.50000 | 9 | 0.5327193242 |
| Diane | 1955 | 200 | 1 | 987 | 14730 | 1 | 9.88889 | 10 | 0.9630864089 |
| Ione | 1955 | 7 | 3 | 960 | 6200 | 0 | 5.94444 | 11 | -0.2591568553 |
| Flossy | 1956 | 15 | 2 | 975 | 1540 | 1 | 7.00000 | 12 | 0.0679232445 |
| Helene | 1958 | 1 | 3 | 946 | 540 | 1 | 9.88889 | 13 | 0.9630864089 |
| Debra | 1959 | 0 | 1 | 984 | 430 | 1 | 9.88889 | 14 | 0.9630864089 |
| Gracie | 1959 | 22 | 3 | 950 | 510 | 1 | 9.77778 | 15 | 0.9286574139 |
| Donna | 1960 | 50 | 4 | 930 | 53270 | 1 | 9.27778 | 16 | 0.7737253874 |
| Ethel | 1960 | 0 | 1 | 981 | 35 | 1 | 8.72222 | 17 | 0.6015773141 |
| Carla | 1961 | 46 | 4 | 931 | 15850 | 1 | 9.50000 | 18 | 0.8425833773 |
| Cindy | 1963 | 3 | 1 | 996 | 300 | 1 | 9.94444 | 19 | 0.9802993570 |
| Cleo | 1964 | 3 | 2 | 968 | 6450 | 1 | 7.94444 | 20 | 0.3605712508 |
| Dora | 1964 | 5 | 2 | 966 | 16260 | 1 | 9.33333 | 21 | 0.7909383355 |
| Hilda | 1964 | 37 | 3 | 950 | 2770 | 1 | 8.83333 | 22 | 0.6360063090 |
| Isbell | 1964 | 3 | 2 | 974 | 800 | 1 | 9.44444 | 23 | 0.8253673305 |
| Betsy | 1965 | 75 | 3 | 948 | 20000 | 1 | 8.33333 | 24 | 0.4810742824 |
| Alma | 1966 | 6 | 2 | 982 | 730 | 1 | 8.77778 | 25 | 0.6187933608 |
| Inez | 1966 | 3 | 1 | 983 | 99 | 1 | 8.27778 | 26 | 0.4638613343 |
| Beulah | 1967 | 15 | 3 | 950 | 5060 | 1 | 7.27778 | 27 | 0.1539972812 |
| Gladys | 1968 | 3 | 2 | 977 | 800 | 1 | 8.94444 | 28 | 0.6704353039 |
| Camille | 1969 | 256 | 5 | 909 | 23040 | 1 | 9.05556 | 29 | 0.7048673975 |
| Celia | 1970 | 22 | 3 | 945 | 6870 | 1 | 9.44444 | 30 | 0.8253673305 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| Bertha | 1996 | 8 | 2 | 974 | 700 | 1 | 8.50000 | 63 | 0.5327193 |
| Fran | 1996 | 26 | 3 | 954 | 8260 | 1 | 7.16667 | 64 | 0.1195683 |
| Danny | 1997 | 10 | 1 | 984 | 200 | 0 | 2.22222 | 65 | -1.4125390 |
| Bonnie | 1998 | 3 | 2 | 964 | 1650 | 1 | 9.38889 | 66 | 0.8081544 |
| Earl | 1998 | 3 | 1 | 987 | 160 | 0 | 1.88889 | 67 | -1.5158260 |
| Georges | 1998 | 1 | 2 | 964 | 3870 | 0 | 2.27778 | 68 | -1.3953230 |
| Bret | 1999 | 0 | 3 | 951 | 94 | 0 | 2.33333 | 69 | -1.3781100 |
| Floyd | 1999 | 56 | 2 | 956 | 8130 | 0 | 1.83333 | 70 | -1.5330421 |
| Irene | 1999 | 8 | 1 | 964 | 1430 | 1 | 9.27778 | 71 | 0.7737254 |
| Lili | 2002 | 2 | 1 | 963 | 1260 | 1 | 10.33333 | 72 | 1.1008024 |
| Claudette | 2003 | 3 | 1 | 979 | 250 | 1 | 9.16667 | 73 | 0.7392964 |
| Isabel | 2003 | 51 | 2 | 957 | 4980 | 1 | 9.38889 | 74 | 0.8081544 |
| Alex | 2004 | 1 | 1 | 972 | 5 | 0 | 4.16667 | 75 | -0.8100239 |
| Charley | 2004 | 10 | 4 | 941 | 20510 | 0 | 2.88889 | 76 | -1.2059620 |
| Frances | 2004 | 7 | 2 | 960 | 12620 | 1 | 6.00000 | 77 | -0.2419408 |
| Gaston | 2004 | 8 | 1 | 985 | 170 | 0 | 2.66667 | 78 | -1.2748200 |
| Ivan | 2004 | 25 | 3 | 946 | 18590 | 0 | 1.05556 | 79 | -1.7740450 |
| Jeanne | 2004 | 5 | 3 | 950 | 10210 | 1 | 8.50000 | 80 | 0.5327193 |
| Cindy | 2005 | 1 | 1 | 991 | 350 | 1 | 9.94444 | 81 | 0.9802994 |
| Dennis | 2005 | 15 | 3 | 946 | 2650 | 0 | 2.44444 | 82 | -1.3436810 |
| Ophelia | 2005 | 1 | 1 | 982 | 91 | 1 | 9.16667 | 83 | 0.7392964 |
| Rita | 2005 | 62 | 3 | 937 | 10690 | 1 | 9.50000 | 84 | 0.8425834 |
| Wilma | 2005 | 5 | 3 | 950 | 25960 | 1 | 8.61111 | 85 | 0.5671483 |
| Humberto | 2007 | 1 | 1 | 985 | 51 | 0 | 2.38889 | 86 | -1.3608940 |
| Dolly | 2008 | 1 | 1 | 967 | 1110 | 1 | 9.83333 | 87 | 0.9458704 |
| Gustav | 2008 | 52 | 2 | 954 | 4360 | 0 | 1.72222 | 88 | -1.5674711 |
| Ike | 2008 | 84 | 2 | 950 | 20370 | 0 | 1.88889 | 89 | -1.5158260 |
| Irene | 2011 | 41 | 1 | 952 | 7110 | 1 | 9.27778 | 90 | 0.7737254 |
| Isaac | 2012 | 5 | 1 | 966 | 24000 | 0 | 1.94444 | 91 | -1.4986131 |
| Sandy | 2012 | 159 | 2 | 942 | 75000 | 1 | 9.00000 | 92 | 0.6876514 |
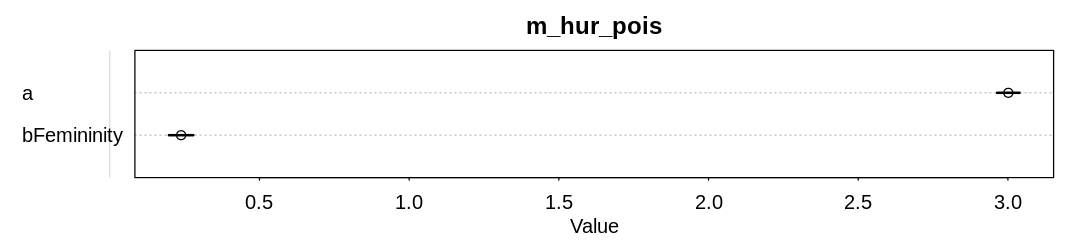
The model with the femininity predictor appears to have inferred a minor but significant association of deaths with femininity:
iplot(function() {
plot(precis(m_hur_pois), main="m_hur_pois")
}, ar=4.5)
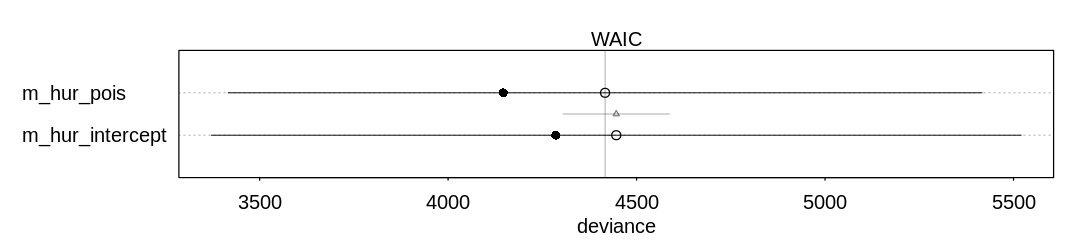
The model with the femininity predictor performs only marginally better on WAIC than the intercept-only model:
iplot(function() {
plot(compare(m_hur_pois, m_hur_intercept))
}, ar=4.5)
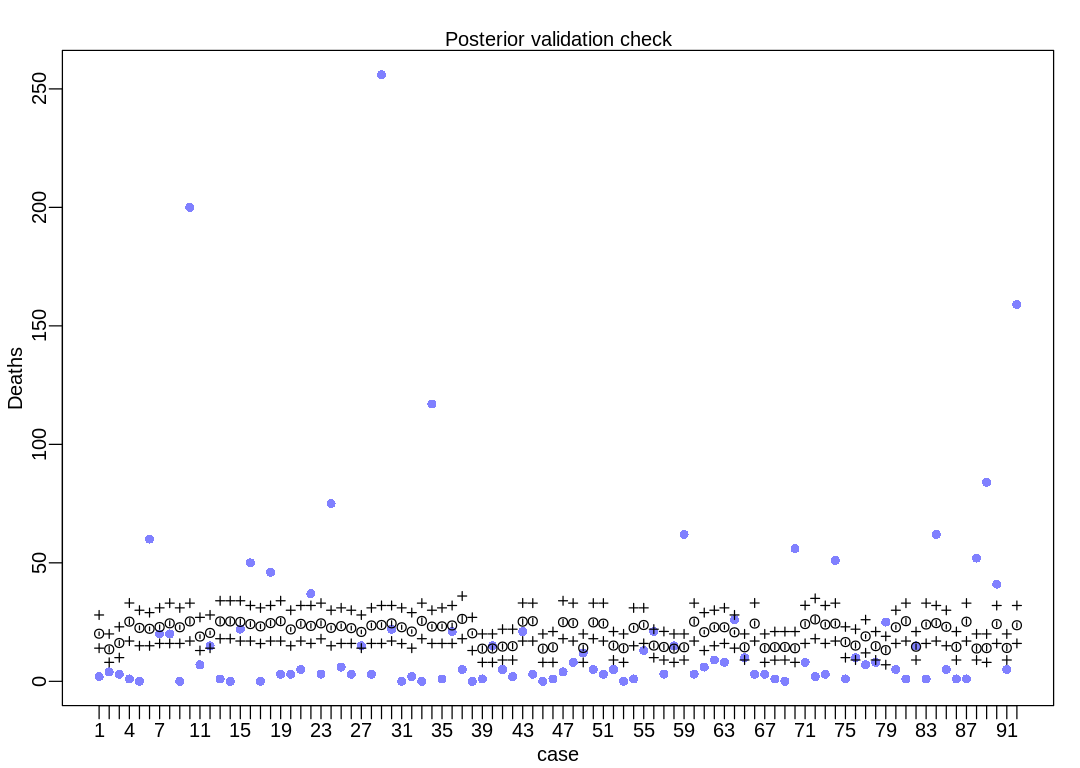
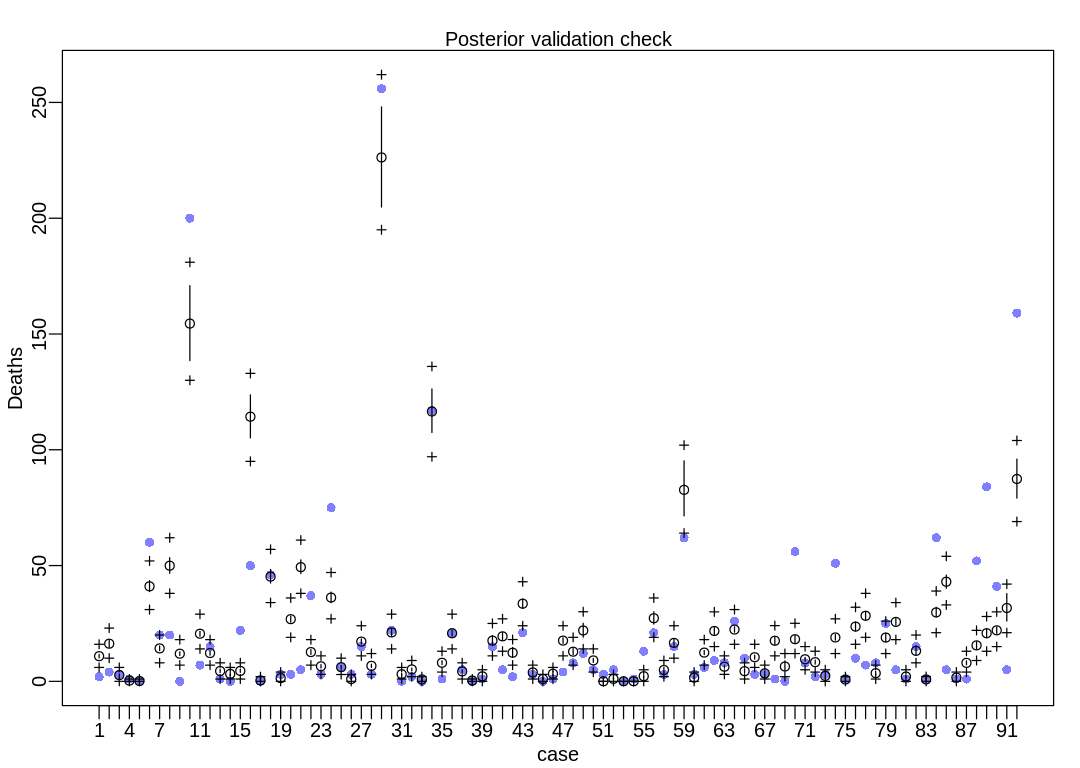
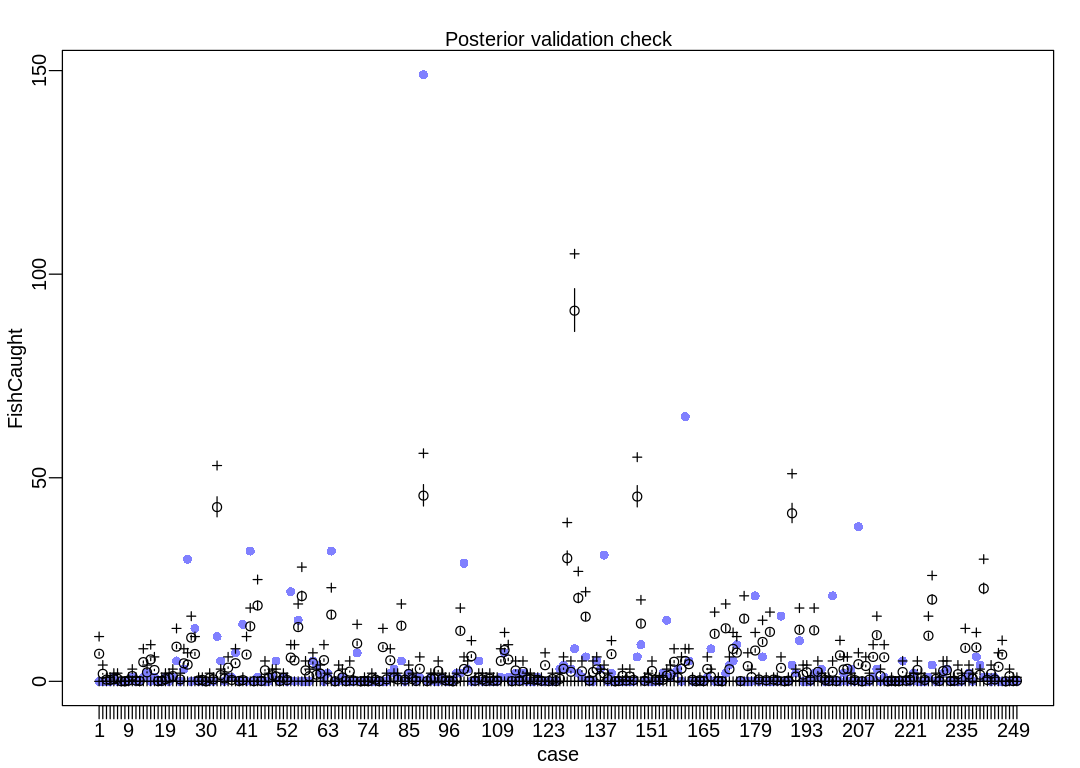
The model is not accurate when it attempts to retrodict the death toll on storms with many deaths. The predictions it makes are less dispersed than the data, that is, the data are overdispersed relative to the predictions (Overdispersion).
iplot(function() {
result <- postcheck(m_hur_pois, window=92)
display_markdown("The raw data:")
display(result)
})
The raw data:
- $mean
-
- 20.0909638320166
- 13.5143028318176
- 16.1733109878181
- 25.1787772660708
- 22.5358282653805
- 22.1689133858378
- 22.908911670051
- 24.464810148839
- 22.8150541596038
- 25.2824892298876
- 18.8935278512169
- 20.4230650001127
- 25.2824892298876
- 25.2824892298876
- 25.0755146831786
- 24.1651462880186
- 23.1928029015675
- 24.5655442972114
- 25.3866150597648
- 21.8976943074182
- 24.2646129731868
- 23.3840464063741
- 24.464810148839
- 22.5358282653805
- 23.2882344448313
- 22.4435351630679
- 20.8460304273314
- 23.5768871486183
- 23.7713562060926
- 24.464810148839
- 22.8150541596038
- 21.0176877923093
- 25.4911940714126
- 23.0977844592675
- 23.1928029015675
- 23.5768871486183
- 26.3436111294371
- 20.256325111861
- 13.7926856723714
- 13.8490494913378
- 14.6631794659105
- 14.8438538180013
- 25.1787772660708
- 25.3866150597648
- 13.7926856723714
- 14.366994702575
- 24.972662491807
- 24.5655442972114
- 14.0768685409229
- 24.8702559127626
- 24.3645123304141
- 15.0882588251682
- 13.9624883489791
- 22.5358282653805
- 23.7713562060926
- 15.0882588251682
- 14.4847323337607
- 13.7926856723714
- 14.366994702575
- 25.1787772660708
- 20.7607272428806
- 22.8150541596038
- 22.8150541596038
- 20.6757928318801
- 14.366994702575
- 24.3645123304141
- 14.0195654325719
- 14.425747144927
- 14.4847323337607
- 13.9624883489791
- 24.1651462880186
- 26.1278374211388
- 23.9674321978027
- 24.3645123304141
- 16.5748764082044
- 15.0882588251682
- 18.9710650264949
- 14.8438538180013
- 13.1877497964257
- 22.8150541596038
- 25.3866150597648
- 14.6034474230733
- 23.9674321978027
- 24.5655442972114
- 23.0031432446451
- 14.5439725264936
- 25.1787772660708
- 13.8490494913378
- 14.0195654325719
- 24.1651462880186
- 14.0768685409229
- 23.6739194496856
- $PI
A matrix: 2 × 92 of type dbl 5% 19.36255 12.35423 15.17837 24.04287 21.72629 21.38360 22.05466 23.41482 21.96716 24.12438 ⋯ 23.00362 23.51569 22.14179 13.44805 24.04287 12.71166 12.88551 23.18141 12.94499 22.72557 94% 20.82471 14.72758 17.18187 26.33569 23.37489 22.96874 23.77376 25.52207 23.66849 26.46576 ⋯ 24.93669 25.63526 23.88443 15.68156 26.33569 15.04457 15.19707 25.16973 15.24818 24.61595 - $outPI
A matrix: 2 × 92 of type dbl 5% 14 8 10 17.000 15 15 16 16 16 17 ⋯ 16 17 15 9 17 9 8 16 9 16 94% 28 20 23 33.055 30 29 31 33 31 33 ⋯ 33 32 30 21 33 20 20 32 20 32
12H2. Counts are nearly always over-dispersed relative to Poisson. So fit a gamma-Poisson (aka
negative-binomial) model to predict deaths using femininity. Show that the over-dispersed model
no longer shows as precise a positive association between femininity and deaths, with an 89% interval
that overlaps zero. Can you explain why the association diminished in strength?
data(Hurricanes)
hur <- Hurricanes
hur$index = 1:92
hur$StdFem = standardize(hur$femininity)
hur_dat = list(
Deaths = hur$deaths,
Femininity = hur$StdFem
)
m_hur_gamma_pois <- map(
alist(
Deaths ~ dgampois(lambda, phi),
log(lambda) <- a + bFemininity * Femininity,
a ~ dnorm(3, 0.5),
bFemininity ~ dnorm(0, 1.0),
phi ~ dexp(1)
),
data = hur_dat
)
Answer. The model no longer infers an association of deaths with femininity:
iplot(function() {
plot(precis(m_hur_gamma_pois), main="m_hur_gamma_pois")
}, ar=4.5)
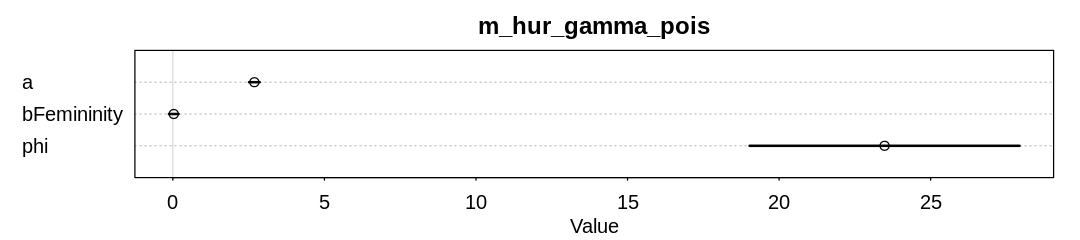
The association has diminished in strength because the influence of outliers are diminished. Notice the storms with the greatest death toll are female in the original data: Diane, Camille, and Sandy.
12H3. In the data, there are two measures of a hurricane’s potential to cause death:
damage_norm and min_pressure. Consult ?Hurricanes for their meanings. It makes some sense to
imagine that femininity of a name matters more when the hurricane is itself deadly. This implies an
interaction between femininity and either or both of damage_norm and min_pressure. Fit a series
of models evaluating these interactions. Interpret and compare the models. In interpreting the
estimates, it may help to generate counterfactual predictions contrasting hurricanes with masculine
and feminine names. Are the effect sizes plausible?
data(Hurricanes)
hur <- Hurricanes
hur$index = 1:92
hur$StdFem = standardize(hur$femininity)
hur$StdDam = standardize(hur$damage_norm)
hur$StdPres = standardize(hur$min_pressure)
Answer. To get some unreasonable answers, lets go back to the Poisson distribution rather than the Gamma-Poisson. It’s likely this is what the authors of the original paper were using.
hur_dat = list(
Deaths = hur$deaths,
StdFem = hur$StdFem,
StdDam = hur$StdDam
)
m_fem_dam_inter <- quap(
alist(
Deaths ~ dpois(lambda),
log(lambda) <- a + bFem * StdFem + bDam * StdDam + bFemDamInter * StdFem * StdDam,
a ~ dnorm(3, 0.5),
bFem ~ dnorm(0, 1.0),
bDam ~ dnorm(0, 1.0),
bFemDamInter ~ dnorm(0, 0.4)
),
data = hur_dat
)
In the first model, lets assume an interaction between femininity and damage_norm. Damage is
unsurprisingly associated with more deaths. The model also believes there is a minor interaction
between damage and femininity, where together they increase deaths more than either alone.
iplot(function() {
plot(precis(m_fem_dam_inter), main="m_fem_dam_inter")
}, ar=4.5)
hur_dat = list(
Deaths = hur$deaths,
StdFem = hur$StdFem,
StdPres = hur$StdPres
)
m_fem_pres_inter <- quap(
alist(
Deaths ~ dpois(lambda),
log(lambda) <- a + bFem * StdFem + bPres * StdPres + bFemPresInter * StdFem * StdPres,
a ~ dnorm(3, 0.5),
bFem ~ dnorm(0, 1.0),
bPres ~ dnorm(0, 1.0),
bFemPresInter ~ dnorm(0, 0.4)
),
data = hur_dat
)
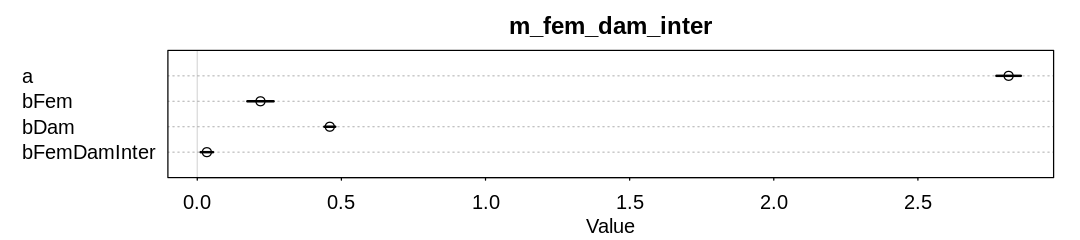
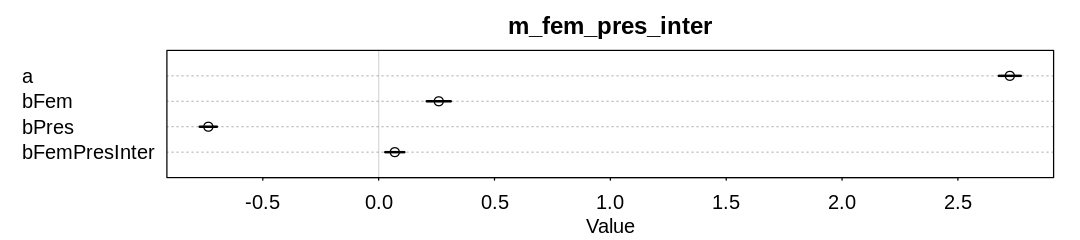
In the second model, lets assume an interaction between femininity and min_pressure. Pressure is
unsurprisingly negatively associated with more deaths; lower pressure indicates a stronger storm.
There also appears to be a minor interaction between pressure and femininity, though now the
association is in the opposite direction. That is, when both pressure and femininity are positive,
which means the storm is weak and feminine, more deaths occur.
iplot(function() {
plot(precis(m_fem_pres_inter), main="m_fem_pres_inter")
}, ar=4.5)
hur_dat = list(
Deaths = hur$deaths,
StdFem = hur$StdFem,
StdPres = hur$StdPres,
StdDam = hur$StdDam
)
m_fem_dam_pres_inter <- quap(
alist(
Deaths ~ dpois(lambda),
log(lambda) <- a + bFem * StdFem + bPres * StdPres + bDam * StdDam +
bFemPresInter * StdFem * StdPres +
bFemDamInter * StdFem * StdDam +
bDamPresInter * StdDam * StdPres +
bFDPInter * StdPres * StdDam * StdPres,
a ~ dnorm(3, 0.5),
bFem ~ dnorm(0, 1.0),
bDam ~ dnorm(0, 1.0),
bPres ~ dnorm(0, 1.0),
bFemPresInter ~ dnorm(0, 0.4),
bFemDamInter ~ dnorm(0, 0.4),
bDamPresInter ~ dnorm(0, 0.4),
bFDPInter ~ dnorm(0, 0.4)
),
data = hur_dat
)
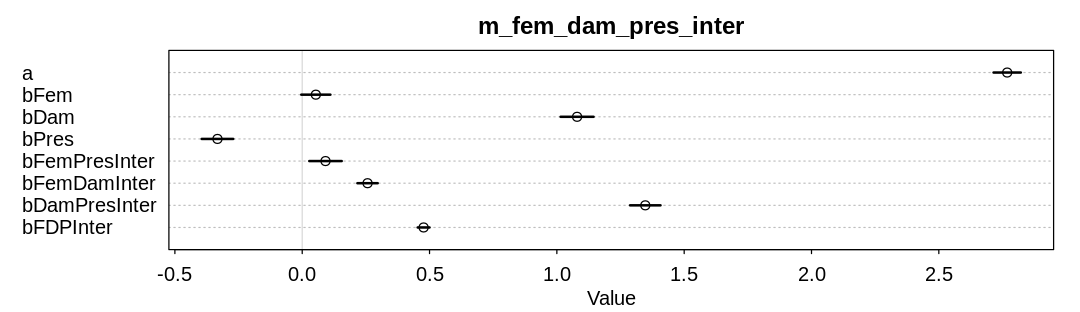
In the third model, lets assume interactions between all three predictors. The influence of femininity has dropped significantly. The three-way interaction term is hard to interpret on the parameter scale. Lets try to interpret it on the outcome scale.
iplot(function() {
plot(precis(m_fem_dam_pres_inter), main="m_fem_dam_pres_inter")
}, ar=3.5)
To some extent this is a counterfactual plot rather than just a posterior prediction plot because we’re considering extremes of at least some predictors. Still, many of the predictors vary here only within the range of the data.
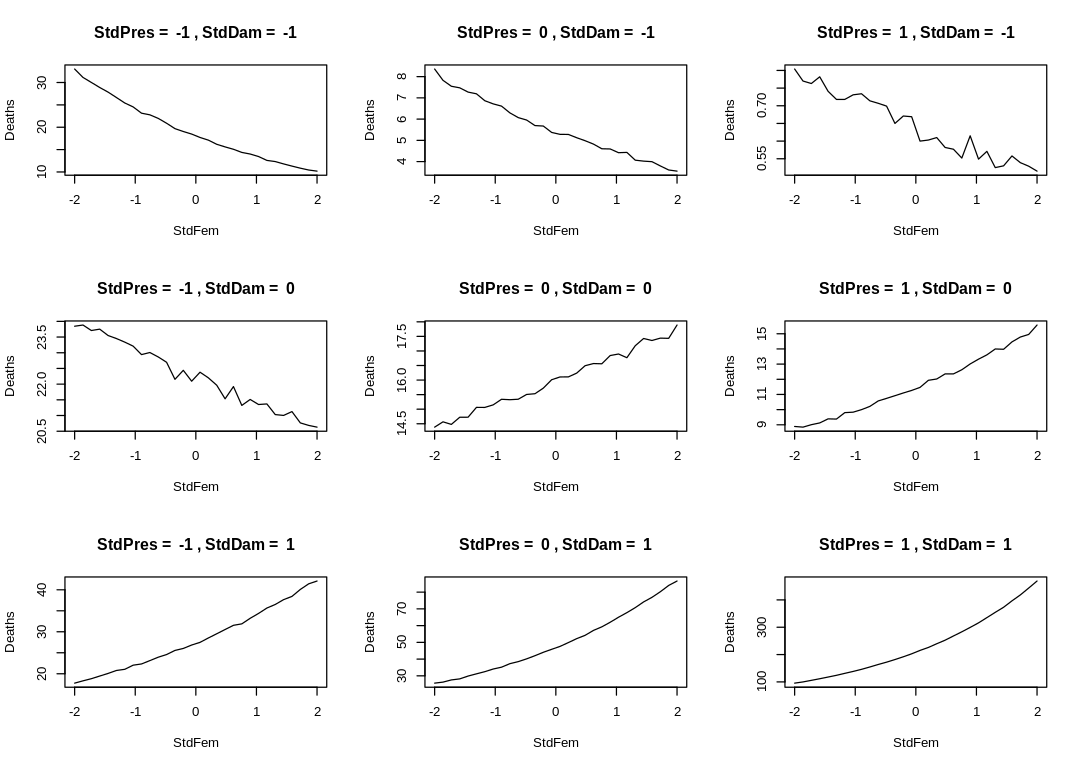
In the following Enneaptych, we hold damage and pressure at typical values and vary femininity. We observe the effect on deaths. Many of the effect sizes are inplausible. In the bottom-right plot, notice an implication of the non-negative three-way interaction parameter: when pressure is high and damage is low femininity has a dramatic effect on deaths.
iplot(function() {
par(mfrow=c(3,3))
for (d in -1: 1) {
for (p in -1:1) {
sim_dat = data.frame(StdPres=p, StdDam=d, StdFem=seq(from=-2, to=2, length.out=30))
s <- sim(m_fem_dam_pres_inter, data=sim_dat)
plot(sim_dat$StdFem, colMeans(s),
main=paste("StdPres = ", p, ",", "StdDam = ", d),
type="l", ylab="Deaths", xlab="StdFem")
}
}
})
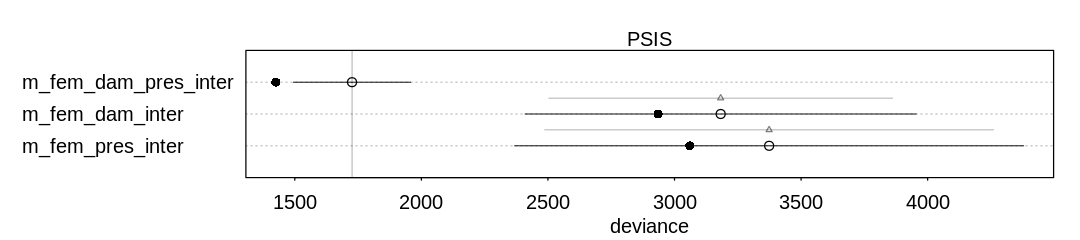
Ignoring the known outliers, PSIS believes the full interaction model performs best:
iplot(function() {
plot(compare(m_fem_dam_pres_inter, m_fem_pres_inter, m_fem_dam_inter, func=PSIS))
}, ar=4.5)
Some Pareto k values are very high (>1). Set pointwise=TRUE to inspect individual points.
Some Pareto k values are very high (>1). Set pointwise=TRUE to inspect individual points.
Some Pareto k values are very high (>1). Set pointwise=TRUE to inspect individual points.
12H4. In the original hurricanes paper, storm damage (damage_norm) was used directly. This
assumption implies that mortality increases exponentially with a linear increase in storm strength,
because a Poisson regression uses a log link. So it’s worth exploring an alternative hypothesis:
that the logarithm of storm strength is what matters. Explore this by using the logarithm of
damage_norm as a predictor. Using the best model structure from the previous problem, compare a
model that uses log(damage_norm) to a model that uses damage_norm directly. Compare their
PSIS/WAIC values as well as their implied predictions. What do you conclude?
Answer We’ll use the full interaction model as our baseline, though it probably doesn’t matter which model we start from.
data(Hurricanes)
hur <- Hurricanes
hur$index = 1:92
hur$StdFem = standardize(hur$femininity)
hur$StdDam = standardize(hur$damage_norm)
hur$StdLogDam = standardize(log(hur$damage_norm))
hur$StdPres = standardize(hur$min_pressure)
hur_dat = list(
Deaths = hur$deaths,
StdFem = hur$StdFem,
StdPres = hur$StdPres,
StdDam = hur$StdDam
)
m_fem_dam_pres_inter <- quap(
alist(
Deaths ~ dpois(lambda),
log(lambda) <- a + bFem * StdFem + bPres * StdPres + bDam * StdDam +
bFemPresInter * StdFem * StdPres +
bFemDamInter * StdFem * StdDam +
bDamPresInter * StdDam * StdPres +
bFDPInter * StdPres * StdDam * StdPres,
a ~ dnorm(3, 0.5),
bFem ~ dnorm(0, 1.0),
bDam ~ dnorm(0, 1.0),
bPres ~ dnorm(0, 1.0),
bFemPresInter ~ dnorm(0, 0.4),
bFemDamInter ~ dnorm(0, 0.4),
bDamPresInter ~ dnorm(0, 0.4),
bFDPInter ~ dnorm(0, 0.4)
),
data = hur_dat
)
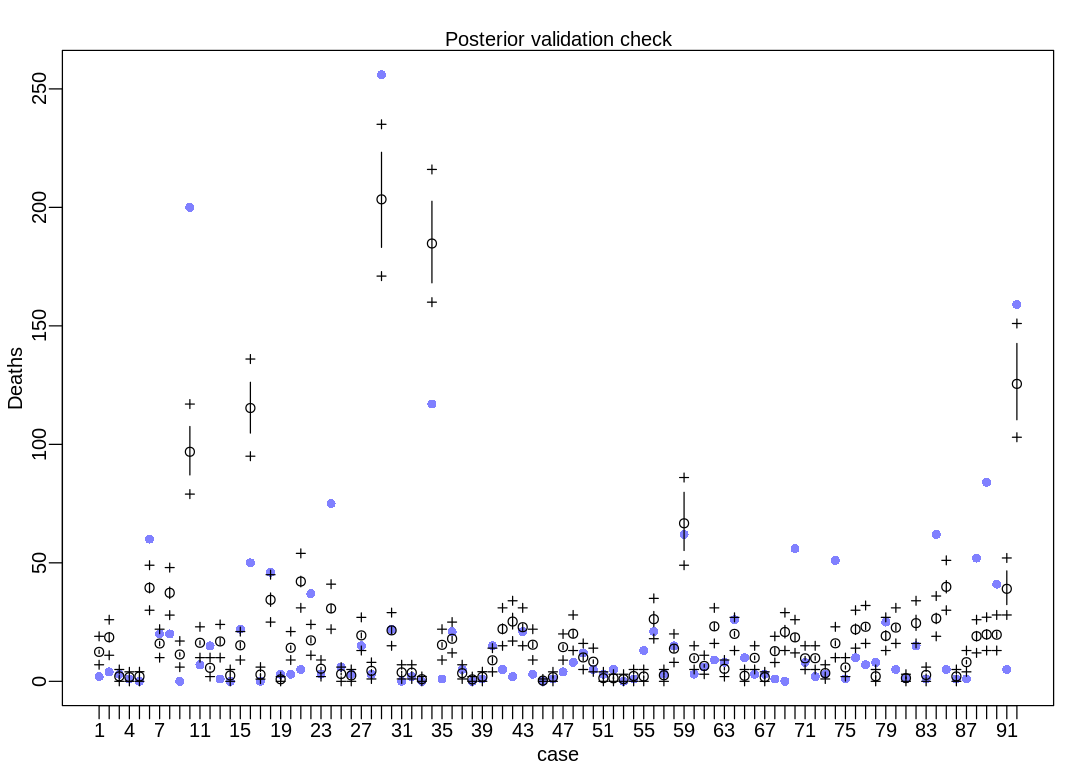
First we’ll run postcheck on the baseline model. Notice there is more dispersion than in the plain
old Poisson model in 12H1:
iplot(function() {
result <- postcheck(m_fem_dam_pres_inter, window=92)
display_markdown("The raw data:")
display(result)
})
hur_dat = list(
Deaths = hur$deaths,
StdFem = hur$StdFem,
StdPres = hur$StdPres,
StdLogDam = hur$StdLogDam
)
m_fem_logdam_pres_inter <- quap(
alist(
Deaths ~ dpois(lambda),
log(lambda) <- a + bFem * StdFem + bPres * StdPres + bDam * StdLogDam +
bFemPresInter * StdFem * StdPres +
bFemDamInter * StdFem * StdLogDam +
bDamPresInter * StdLogDam * StdPres +
bFDPInter * StdPres * StdLogDam * StdPres,
a ~ dnorm(3, 0.5),
bFem ~ dnorm(0, 1.0),
bDam ~ dnorm(0, 1.0),
bPres ~ dnorm(0, 1.0),
bFemPresInter ~ dnorm(0, 0.4),
bFemDamInter ~ dnorm(0, 0.4),
bDamPresInter ~ dnorm(0, 0.4),
bFDPInter ~ dnorm(0, 0.4)
),
data = hur_dat
)
The raw data:
- $mean
-
- 12.4192462885702
- 18.5938168455421
- 2.11381194508373
- 1.77493954809473
- 2.06508536687668
- 39.4690667431158
- 15.9610663893155
- 37.358298332016
- 11.3212801161669
- 96.8666494831281
- 16.2604276543812
- 5.69564845261434
- 16.8378743556131
- 2.48682003620383
- 15.2026808132042
- 115.329975983529
- 2.86569503514338
- 34.4103632635181
- 0.858249074601839
- 14.1553490927226
- 42.1341677686966
- 17.2845656286884
- 5.31305551387514
- 30.7520944493205
- 3.0837990192428
- 2.48299529863905
- 19.392170532057
- 4.41411579164916
- 203.384909416927
- 21.504460642179
- 3.7775803550042
- 3.66808922049433
- 0.910404527206778
- 184.758064484002
- 15.4002185551508
- 17.9602111705932
- 3.43535493602018
- 0.82149797513125
- 1.88977382488604
- 8.87356128718296
- 22.0901686916238
- 25.2305705523166
- 22.8483178647742
- 15.4562893566688
- 0.314647353158313
- 1.75574042405943
- 14.4320375051793
- 20.0984933373014
- 10.0849844222571
- 8.29586575764401
- 1.34149367017358
- 1.28626217978254
- 0.923591644163742
- 2.23901284894469
- 2.0416393055465
- 26.2255624651538
- 2.56590615956333
- 13.8000154129816
- 66.6899024324415
- 9.76544195918975
- 6.49516889581563
- 23.198571097516
- 5.28000404637473
- 20.0148275741584
- 2.33439640074048
- 9.90300242282889
- 1.7537386186061
- 12.7469121252857
- 20.8265191297282
- 18.5405774511215
- 9.72158475950537
- 9.76524130280055
- 3.46925119527546
- 16.0444508846542
- 5.8631354723675
- 21.9122731810267
- 23.0232995511413
- 2.12003366427668
- 19.1718685965319
- 22.6813508455968
- 1.37381187319183
- 24.5510734365911
- 2.68041577016984
- 26.5396233012641
- 39.8759313082704
- 2.0679848610754
- 8.08639310904994
- 19.0363742456932
- 19.7655928225731
- 19.6766678257971
- 39.04416705125
- 125.520453680288
- $PI
A matrix: 2 × 92 of type dbl 5% 11.70273 16.68520 1.759826 1.509584 1.797090 37.32644 15.03229 34.91078 10.57674 87.09672 ⋯ 2.347731 24.34544 37.31906 1.655340 7.358798 17.08223 17.76707 18.50674 32.38596 110.3816 94% 13.16521 20.59954 2.546389 2.081431 2.374178 41.58615 16.85816 39.80710 12.07033 107.53285 ⋯ 3.058250 28.75276 42.51885 2.554984 8.830958 21.07172 21.80052 20.90788 46.59840 142.5699 - $outPI
A matrix: 2 × 92 of type dbl 5% 7 11 0 0 0.000 30 10 27.945 6 79 ⋯ 0 19 30.000 0 4 12 13.000 13 28.000 103 94% 19 26 5 4 4.055 49 22 48.000 17 117 ⋯ 6 36 51.055 5 13 26 27.055 28 52.055 151
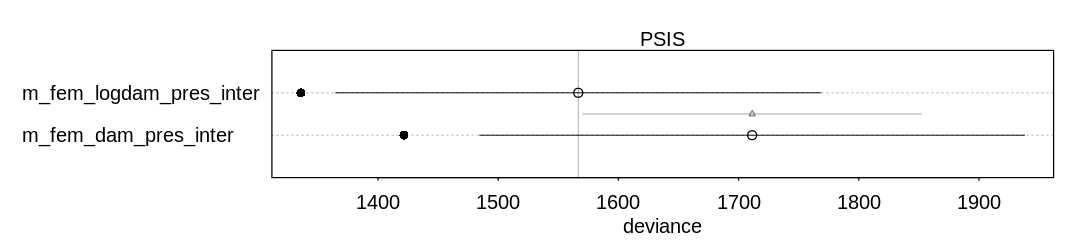
Ignoring the known outliers, comparing the models using PSIS, it appears the log(damage) model
improves performance:
iplot(function() {
plot(compare(m_fem_dam_pres_inter, m_fem_logdam_pres_inter, func=PSIS))
}, ar=4.5)
Some Pareto k values are very high (>1). Set pointwise=TRUE to inspect individual points.
Some Pareto k values are very high (>1). Set pointwise=TRUE to inspect individual points.
Run postcheck to visualize how much the model has changed on the outcome scale:
iplot(function() {
result <- postcheck(m_fem_logdam_pres_inter, window=92)
display_markdown("The raw data:")
display(result)
})
The raw data:
- $mean
-
- 10.8699882836877
- 16.2595681620299
- 2.67255400951333
- 0.324891055351021
- 0.136794243571386
- 41.0238249932624
- 14.2206464899141
- 49.9349468125643
- 11.9320898678004
- 154.499154480515
- 20.5619727279893
- 12.2148037384162
- 4.4561389708142
- 3.19519090024716
- 4.50855930854229
- 114.29724808675
- 0.399705113340796
- 45.2297046566204
- 1.39761631106256
- 26.8534972701785
- 49.3207414632433
- 12.6381635360833
- 6.49466550321014
- 36.1755561834113
- 6.11465018583331
- 0.988682407787108
- 17.1613394443972
- 6.69589713527632
- 226.252793873048
- 21.1241232209717
- 3.07289094570769
- 5.15026540733052
- 0.832841989107325
- 116.526910474906
- 8.02121391702632
- 20.756373368377
- 4.27972358733588
- 0.339424235355401
- 2.15575129089143
- 17.5665174746855
- 19.4286826354081
- 12.3566344076102
- 33.532691484247
- 3.85241425339933
- 0.989993520191923
- 3.27361461134978
- 17.5895153295144
- 12.8045063246198
- 21.9652390357988
- 9.03333226982972
- 0.0193755906404282
- 1.05886140918945
- 0.0280480135459265
- 0.0432815209135604
- 2.21005348131204
- 27.2597115785741
- 4.89900537748048
- 16.6009046234761
- 82.6953189366748
- 1.59268707136014
- 12.4127603882858
- 21.7426511119712
- 6.3025973303236
- 22.3686319891363
- 4.3810954541357
- 10.4123953524991
- 3.45049157776192
- 17.534453890374
- 6.39968459072767
- 18.1958495648466
- 9.53385569354962
- 8.27301885619899
- 2.35454398034847
- 18.9537906368423
- 0.78951448157749
- 23.6402882413444
- 28.3081191196519
- 3.51651372895571
- 18.906859578512
- 25.6698728834182
- 2.07834564607815
- 13.1695536352315
- 0.824403690257513
- 29.7669102790378
- 43.0102158772508
- 1.61453240811467
- 7.97032785414566
- 15.5282386088827
- 20.7863785281744
- 22.0236036269154
- 31.6724471286218
- 87.3912071579769
- $PI
A matrix: 2 × 92 of type dbl 5% 10.10567 14.67018 2.150698 0.2387359 0.09846129 38.75203 13.26998 46.65318 11.14271 138.5229 ⋯ 0.6540991 27.78764 40.18345 1.192299 7.169494 13.90945 18.80923 20.68352 26.04830 79.13909 94% 11.64052 17.90678 3.232551 0.4271335 0.18349747 43.48464 15.25742 53.38582 12.77133 170.8846 ⋯ 1.0130670 32.00657 45.90527 2.135289 8.794958 17.12137 22.80798 23.46071 37.90813 95.99470 - $outPI
A matrix: 2 × 92 of type dbl 5% 6 10 0 0 0 31 8 38 7 130 ⋯ 0.000 21 33 0 4 9 13 15 21 69 94% 16 23 6 1 1 52 20 62 18 181 ⋯ 2.055 39 54 4 13 22 28 30 42 104
12H5. One hypothesis from developmental psychology, usually attributed to Carol Gilligan,
proposes that women and men have different average tendencies in moral reasoning. Like most
hypotheses in social psychology, it is merely descriptive, not causal. The notion is that women are
more concerned with care (avoiding harm), while men are more concerned with justice and rights.
Evaluate this hypothesis, using the Trolley data, supposing that contact provides a proxy for
physical harm. Are women more or less bothered by contact than are men, in these data? Figure out
the model(s) that is needed to address this question.
Answer. The head of the Trolley data.frame:
data(Trolley)
d <- Trolley
display(head(Trolley))
| case | response | order | id | age | male | edu | action | intention | contact | story | action2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <fct> | <int> | <int> | <fct> | <int> | <int> | <fct> | <int> | <int> | <int> | <fct> | <int> | |
| 1 | cfaqu | 4 | 2 | 96;434 | 14 | 0 | Middle School | 0 | 0 | 1 | aqu | 1 |
| 2 | cfbur | 3 | 31 | 96;434 | 14 | 0 | Middle School | 0 | 0 | 1 | bur | 1 |
| 3 | cfrub | 4 | 16 | 96;434 | 14 | 0 | Middle School | 0 | 0 | 1 | rub | 1 |
| 4 | cibox | 3 | 32 | 96;434 | 14 | 0 | Middle School | 0 | 1 | 1 | box | 1 |
| 5 | cibur | 3 | 4 | 96;434 | 14 | 0 | Middle School | 0 | 1 | 1 | bur | 1 |
| 6 | cispe | 3 | 9 | 96;434 | 14 | 0 | Middle School | 0 | 1 | 1 | spe | 1 |
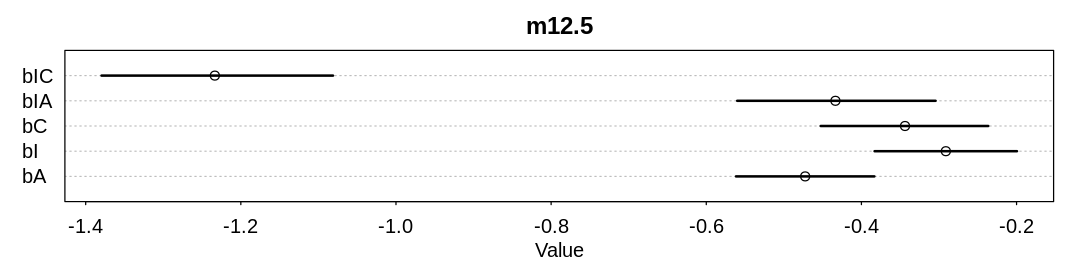
We’ll start from model m12.5 in the text, converting bC and bIC to index variables based on
gender. Before doing the conversion, lets reproduce the precis summary for m12.5 in the text:
dat <- list(
R = d$response,
A = d$action,
I = d$intention,
C = d$contact
)
m12.5 <- ulam(
alist(
R ~ dordlogit(phi, cutpoints),
phi <- bA * A + bC * C + BI * I,
BI <- bI + bIA * A + bIC * C,
c(bA, bI, bC, bIA, bIC) ~ dnorm(0, 0.5),
cutpoints ~ dnorm(0, 1.5)
),
data = dat, chains = 4, cores = min(detectCores(), 4), log_lik=TRUE
)
iplot(function() {
plot(precis(m12.5), main="m12.5")
}, ar=4)
dat <- list(
R = d$response,
A = d$action,
I = d$intention,
C = d$contact,
gid = ifelse(d$male == 1, 2, 1)
)
m_trolley_care <- ulam(
alist(
R ~ dordlogit(phi, cutpoints),
phi <- bA * A + bC[gid] * C + BI * I,
BI <- bI + bIA * A + bIC[gid] * C,
c(bA, bI, bIA) ~ dnorm(0, 0.5),
bC[gid] ~ dnorm(0, 0.5),
bIC[gid] ~ dnorm(0, 0.5),
cutpoints ~ dnorm(0, 1.5)
),
data = dat, chains = 4, cores = min(detectCores(), 4), log_lik=TRUE
)
flush.console()
6 vector or matrix parameters hidden. Use depth=2 to show them.
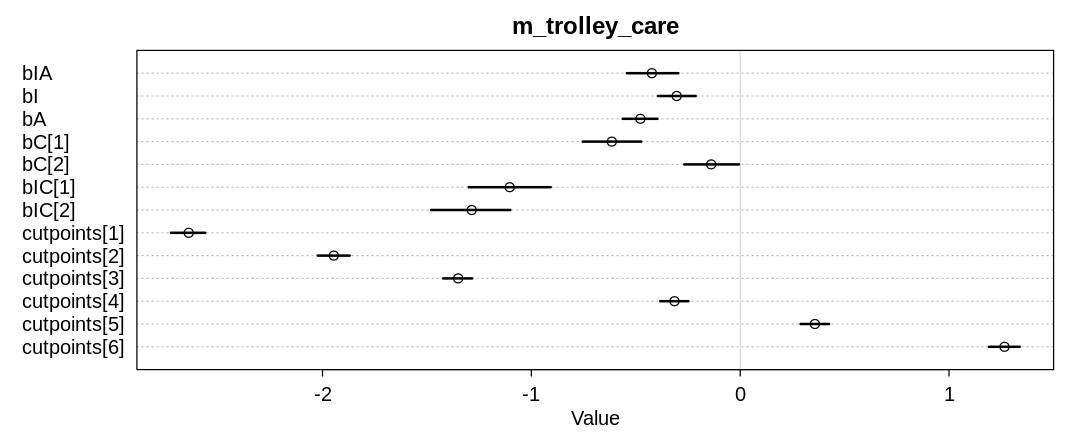
The following plot shows the same precis summary after converting bC and bIC to index
variables based on gender. It appears women disapprove of contact more than men from this data and
model, lending some support to the theory:
iplot(function() {
plot(precis(m_trolley_care, depth=2), main="m_trolley_care")
}, ar=2.5)
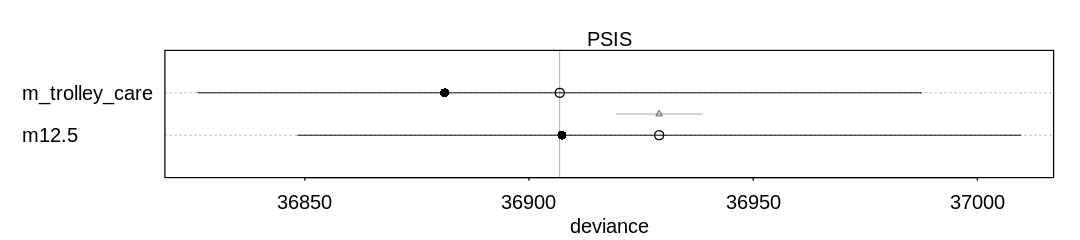
Comparing the models using PSIS, to check for overfitting or outliers:
iplot(function() {
plot(compare(m12.5, m_trolley_care, func=PSIS))
}, ar=4.5)
12H6. The data in data(Fish) are records of visits to a national park. See ?Fish for
details. The question of interest is how many fish an average visitor takes per hour, when fishing.
The problem is that not everyone tried to fish, so the fish_caught numbers are zero-inflated. As
with the monks example in the chapter, there is a process that determines who is fishing (working)
and another process that determines fish per hour (manuscripts per day), conditional on fishing
(working). We want to model both. Otherwise we’ll end up with an underestimate of rate of fish
extraction from the park.
You will model these data using zero-inflated Poisson GLMs. Predict fish_caught as a function
of any of the other variables you think are relevant. One thing you must do, however, is use a
proper Poisson offset/exposure in the Poisson portion of the zero-inflated model. Then use the
hours variable to construct the offset. This will adjust the model for the differing amount of
time individuals spent in the park.
Answer. The short documentation on Fish:
display(help(Fish))
data(Fish)
| Fish {rethinking} | R Documentation |
Fishing data
Description
Fishing data from visitors to a national park.
Usage
data(Fish)
Format
fish_caught : Number of fish caught during visit
livebait : Whether or not group used livebait to fish
camper : Whether or not group had a camper
persons : Number of adults in group
child : Number of children in group
hours : Number of hours group spent in park
The head of Fish:
display(head(Fish))
| fish_caught | livebait | camper | persons | child | hours | |
|---|---|---|---|---|---|---|
| <int> | <int> | <int> | <int> | <int> | <dbl> | |
| 1 | 0 | 0 | 0 | 1 | 0 | 21.124 |
| 2 | 0 | 1 | 1 | 1 | 0 | 5.732 |
| 3 | 0 | 1 | 0 | 1 | 0 | 1.323 |
| 4 | 0 | 1 | 1 | 2 | 1 | 0.548 |
| 5 | 1 | 1 | 0 | 1 | 0 | 1.695 |
| 6 | 0 | 1 | 1 | 4 | 2 | 0.493 |
Notice several outliers in the full dataframe (not represented in head):
display(summary(Fish))
fish_caught livebait camper persons
Min. : 0.000 Min. :0.000 Min. :0.000 Min. :1.000
1st Qu.: 0.000 1st Qu.:1.000 1st Qu.:0.000 1st Qu.:2.000
Median : 0.000 Median :1.000 Median :1.000 Median :2.000
Mean : 3.296 Mean :0.864 Mean :0.588 Mean :2.528
3rd Qu.: 2.000 3rd Qu.:1.000 3rd Qu.:1.000 3rd Qu.:4.000
Max. :149.000 Max. :1.000 Max. :1.000 Max. :4.000
child hours
Min. :0.000 Min. : 0.0040
1st Qu.:0.000 1st Qu.: 0.2865
Median :0.000 Median : 1.8315
Mean :0.684 Mean : 5.5260
3rd Qu.:1.000 3rd Qu.: 7.3427
Max. :3.000 Max. :71.0360
One complication introduced by the question and data is how to infer fish per ‘average visitor’ when
every observation includes several visitors, both adults and children. This solution disregards
children, assuming any fishing they do is offset by the time they take away from fishing. Addressing
persons can be done the same way we address hours, by treating it as an exposure. That is:
$\(
\begin{align}
log \left(\frac{fish}{person \cdot hour}\right) & = log(fish) - log(person \cdot hour) \\
& = log(fish) - log(person) - log(hour)
\end{align}
\)$
fish_dat <- list(
FishCaught = Fish$fish_caught,
LogPersonsInGroup = log(Fish$persons),
LogHours = log(Fish$hours)
)
m_fish <- ulam(
alist(
FishCaught ~ dzipois(p, fish_per_person_hour),
log(fish_per_person_hour) <- LogPersonsInGroup + LogHours + al,
logit(p) <- ap,
ap ~ dnorm(-0.5, 1),
al ~ dnorm(1, 2)
), data = fish_dat, chains = 4
)
flush.console()
SAMPLING FOR MODEL '1a5d1436509eaa3c57b3600d89299fd9' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 0.00012 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 1.2 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 1000 [ 0%] (Warmup)
Chain 1: Iteration: 100 / 1000 [ 10%] (Warmup)
Chain 1: Iteration: 200 / 1000 [ 20%] (Warmup)
Chain 1: Iteration: 300 / 1000 [ 30%] (Warmup)
Chain 1: Iteration: 400 / 1000 [ 40%] (Warmup)
Chain 1: Iteration: 500 / 1000 [ 50%] (Warmup)
Chain 1: Iteration: 501 / 1000 [ 50%] (Sampling)
Chain 1: Iteration: 600 / 1000 [ 60%] (Sampling)
Chain 1: Iteration: 700 / 1000 [ 70%] (Sampling)
Chain 1: Iteration: 800 / 1000 [ 80%] (Sampling)
Chain 1: Iteration: 900 / 1000 [ 90%] (Sampling)
Chain 1: Iteration: 1000 / 1000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 0.427317 seconds (Warm-up)
Chain 1: 0.288993 seconds (Sampling)
Chain 1: 0.71631 seconds (Total)
Chain 1:
SAMPLING FOR MODEL '1a5d1436509eaa3c57b3600d89299fd9' NOW (CHAIN 2).
Chain 2:
Chain 2: Gradient evaluation took 9e-05 seconds
Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.9 seconds.
Chain 2: Adjust your expectations accordingly!
Chain 2:
Chain 2:
Chain 2: Iteration: 1 / 1000 [ 0%] (Warmup)
Chain 2: Iteration: 100 / 1000 [ 10%] (Warmup)
Chain 2: Iteration: 200 / 1000 [ 20%] (Warmup)
Chain 2: Iteration: 300 / 1000 [ 30%] (Warmup)
Chain 2: Iteration: 400 / 1000 [ 40%] (Warmup)
Chain 2: Iteration: 500 / 1000 [ 50%] (Warmup)
Chain 2: Iteration: 501 / 1000 [ 50%] (Sampling)
Chain 2: Iteration: 600 / 1000 [ 60%] (Sampling)
Chain 2: Iteration: 700 / 1000 [ 70%] (Sampling)
Chain 2: Iteration: 800 / 1000 [ 80%] (Sampling)
Chain 2: Iteration: 900 / 1000 [ 90%] (Sampling)
Chain 2: Iteration: 1000 / 1000 [100%] (Sampling)
Chain 2:
Chain 2: Elapsed Time: 0.311079 seconds (Warm-up)
Chain 2: 0.256878 seconds (Sampling)
Chain 2: 0.567957 seconds (Total)
Chain 2:
SAMPLING FOR MODEL '1a5d1436509eaa3c57b3600d89299fd9' NOW (CHAIN 3).
Chain 3:
Chain 3: Gradient evaluation took 9.2e-05 seconds
Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.92 seconds.
Chain 3: Adjust your expectations accordingly!
Chain 3:
Chain 3:
Chain 3: Iteration: 1 / 1000 [ 0%] (Warmup)
Chain 3: Iteration: 100 / 1000 [ 10%] (Warmup)
Chain 3: Iteration: 200 / 1000 [ 20%] (Warmup)
Chain 3: Iteration: 300 / 1000 [ 30%] (Warmup)
Chain 3: Iteration: 400 / 1000 [ 40%] (Warmup)
Chain 3: Iteration: 500 / 1000 [ 50%] (Warmup)
Chain 3: Iteration: 501 / 1000 [ 50%] (Sampling)
Chain 3: Iteration: 600 / 1000 [ 60%] (Sampling)
Chain 3: Iteration: 700 / 1000 [ 70%] (Sampling)
Chain 3: Iteration: 800 / 1000 [ 80%] (Sampling)
Chain 3: Iteration: 900 / 1000 [ 90%] (Sampling)
Chain 3: Iteration: 1000 / 1000 [100%] (Sampling)
Chain 3:
Chain 3: Elapsed Time: 0.348026 seconds (Warm-up)
Chain 3: 0.290844 seconds (Sampling)
Chain 3: 0.63887 seconds (Total)
Chain 3:
SAMPLING FOR MODEL '1a5d1436509eaa3c57b3600d89299fd9' NOW (CHAIN 4).
Chain 4:
Chain 4: Gradient evaluation took 9.7e-05 seconds
Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.97 seconds.
Chain 4: Adjust your expectations accordingly!
Chain 4:
Chain 4:
Chain 4: Iteration: 1 / 1000 [ 0%] (Warmup)
Chain 4: Iteration: 100 / 1000 [ 10%] (Warmup)
Chain 4: Iteration: 200 / 1000 [ 20%] (Warmup)
Chain 4: Iteration: 300 / 1000 [ 30%] (Warmup)
Chain 4: Iteration: 400 / 1000 [ 40%] (Warmup)
Chain 4: Iteration: 500 / 1000 [ 50%] (Warmup)
Chain 4: Iteration: 501 / 1000 [ 50%] (Sampling)
Chain 4: Iteration: 600 / 1000 [ 60%] (Sampling)
Chain 4: Iteration: 700 / 1000 [ 70%] (Sampling)
Chain 4: Iteration: 800 / 1000 [ 80%] (Sampling)
Chain 4: Iteration: 900 / 1000 [ 90%] (Sampling)
Chain 4: Iteration: 1000 / 1000 [100%] (Sampling)
Chain 4:
Chain 4: Elapsed Time: 0.336802 seconds (Warm-up)
Chain 4: 0.291063 seconds (Sampling)
Chain 4: 0.627865 seconds (Total)
Chain 4:
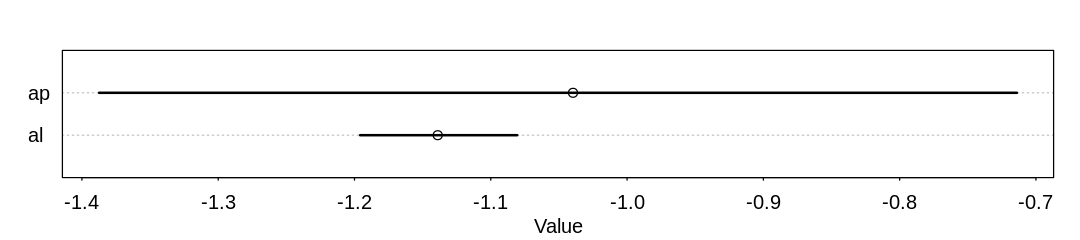
The precision of the parameters, which is rather hard to interpret. We can take from ap that most
visitors who were a part of this survey chose to fish. That is, the probability of not fishing looks
solidly below 0.5, since ap is solidly negative and any score less than zero implies a probability
of less than 0.5:
iplot(function() {
plot(precis(m_fish))
}, ar=4.5)
Finally, a posterior check. Despite the large uncertainty in ap the model is still surprised by
outliers. An obvious alternative based on this chapter is a Gamma-Poisson, but this solution doesn’t
pursue that.
iplot(function() {
result <- postcheck(m_fish, window=250)
display_markdown("The raw data:")
display(result)
})
The raw data:
- $mean
-
- 6.76751586652509
- 1.83636626334604
- 0.423850761759738
- 0.351126556983124
- 0.543028753728462
- 0.6317715058127
- 0.031716723669096
- 0.00512593513843976
- 0.208561485945268
- 1.29141528394067
- 0.185815148768441
- 0.0807334784304262
- 4.7190640368261
- 2.20287062574449
- 5.37806707306176
- 2.77345128084206
- 0.0589482540920572
- 0.136478023060959
- 0.531815770613125
- 0.85346820055022
- 1.28404675217916
- 8.50648936224078
- 0.477352709767203
- 4.4224005406889
- 4.10299070737487
- 10.7221748258314
- 6.71625651514069
- 0.221055952845215
- 0.347282105629294
- 0.0102518702768795
- 0.805412558627347
- 0.494973111805589
- 42.7938695032643
- 1.52881015503966
- 0.693282727473977
- 3.35876899946265
- 0.337030235352414
- 4.4480302163811
- 0.0989946223611178
- 0.293459786675676
- 6.56792476707209
- 13.5132465087118
- 0.0442111905690429
- 18.6173964228132
- 0.0897038649226958
- 2.70425115647313
- 0.868205264073234
- 1.51407309151664
- 1.1994688223949
- 0.0820149622150362
- 0.851545974873305
- 0.31588575290635
- 5.83331418754444
- 5.12657588033206
- 13.3248683923741
- 20.9330376216034
- 2.67605851321171
- 1.23759296498705
- 4.52363775967309
- 1.63357145443152
- 1.89339229176119
- 5.20730935876249
- 0.595249217951317
- 16.3786442510996
- 0.0192222567691491
- 1.58807878007787
- 0.973927676303554
- 0.0605501088228197
- 2.39477282248982
- 0.177485504168477
- 9.29107780936822
- 0.00897038649226958
- 0.0858594135688659
- 0.0384445135382982
- 0.798684768758145
- 0.41648222999823
- 0.0413278520536706
- 8.40781511082581
- 0.504904611136316
- 5.16566113576267
- 0.850905232981
- 0.465178613813408
- 13.6593356601573
- 0.409754440129028
- 1.78702913763856
- 0.555523220628409
- 0.0999557351995753
- 3.07908516347153
- 45.610570861837
- 0.0605501088228197
- 0.650673391635697
- 0.700330888289332
- 2.54022123204305
- 0.656119697720289
- 0.226822629875959
- 0.214328162976012
- 0.0166592891999292
- 0.622801119320431
- 12.402200067455
- 3.0294276668179
- 2.51523229824316
- 6.17867406749682
- 0.161466956860852
- 0.860195990419422
- 0.456208227321139
- 0.0884223811380858
- 0.654838213935679
- 0.140322474414788
- 0.185815148768441
- 4.99906824376337
- 7.68922307860579
- 5.36236889670029
- 0.0509389804382451
- 2.66676775577328
- 0.108605750745692
- 2.33678568123623
- 0.497856450320962
- 0.594608476059012
- 0.411356294859791
- 0.37387289415995
- 0.126866894676384
- 3.96010526539087
- 0.0768890270765964
- 0.574104735505253
- 0.0666371567997169
- 0.609986281474331
- 3.08613332428689
- 30.2356487850331
- 2.34447458394388
- 91.0314821235517
- 20.4697812334669
- 2.47838963943562
- 15.8763026075325
- 0.176844762276172
- 2.28104113660569
- 2.92306451269527
- 1.32249126571746
- 1.93856459516869
- 0.545271350351529
- 6.68421942052545
- 0.0140963216307093
- 0.0967520257380504
- 1.34940242519427
- 0.20760037310681
- 1.18248916224882
- 0.334467267783194
- 45.3811852643918
- 14.1812199314398
- 0.122061330484097
- 0.738134659935325
- 2.54855087664302
- 0.397259973229081
- 0.373232152267645
- 0.393415521875251
- 1.40322474414788
- 1.6249214388854
- 4.80812715985649
- 0.876534908673199
- 3.12361672498673
- 5.00419417890181
- 4.21447979663594
- 0.385406248221439
- 0.0374834006998408
- 0.261422692060428
- 0.0551038027382274
- 3.02173876411024
- 1.18537250076419
- 11.6640654075197
- 0.0970723966842029
- 0.0243481919075889
- 13.0221178482601
- 2.98073128300272
- 7.96442172135078
- 7.05328675049311
- 0.0704816081535467
- 15.3976684139807
- 3.72719558753801
- 1.03960372026481
- 7.5684432319063
- 0.453324888805766
- 9.66302847785125
- 0.278081981260357
- 12.137573665933
- 0.42801558405972
- 0.199911470399151
- 3.3139170670013
- 0.0297944979921811
- 0.172039198083884
- 41.2714667671477
- 1.16615024399505
- 12.6665060980308
- 1.63517330916228
- 2.22529659197516
- 0.370669184698425
- 12.5290669621314
- 2.32204861771321
- 0.872370086373216
- 0.347922847521599
- 0.117896508184114
- 2.34383384205158
- 0.0800927365381213
- 6.38435221492672
- 2.9073663363338
- 3.08260924387921
- 0.538223189536175
- 0.302750544114098
- 4.20967423244365
- 0.0128148378460994
- 3.76980492337629
- 0.414239633375163
- 5.99349966062069
- 11.3372870424441
- 1.3080745731406
- 5.9050772794826
- 0.0115333540614895
- 0.20760037310681
- 0.0525408351690075
- 0.0643945601766495
- 2.27463371768264
- 0.144807667660923
- 0.352408040767733
- 0.965918402649742
- 2.78658648963431
- 0.834886685673376
- 0.329982074537059
- 11.2084979220908
- 20.083413872407
- 0.59044365375903
- 0.0913057196534582
- 2.53413418406616
- 2.72571600986534
- 0.0820149622150361
- 0.161466956860852
- 1.74538091463874
- 0.411997036752096
- 8.19124435122674
- 1.65503630782374
- 0.685593824766318
- 8.32612051955694
- 1.62043624563927
- 22.793752076857
- 0.343437654275464
- 1.84533664983831
- 0.461334162459578
- 3.58206754893093
- 6.54774139746449
- 0.00512593513843976
- 1.26162078594849
- 0.0720834628843091
- 0.149292860907058
- $PI
A matrix: 2 × 250 of type dbl 5% 6.388610 1.733550 0.4001198 0.3314674 0.5126252 0.5963993 0.02994094 0.004838939 0.1968843 1.219110 ⋯ 21.51755 0.3242089 1.742018 0.4355046 3.381511 6.181140 0.004838939 1.190984 0.06804759 0.1409341 94% 7.169384 1.945413 0.4490198 0.3719771 0.5752748 0.6692873 0.03360012 0.005430323 0.2209463 1.368102 ⋯ 24.14729 0.3638316 1.954916 0.4887291 3.794777 6.936559 0.005430323 1.336538 0.07636391 0.1581582 - $outPI
A matrix: 2 × 250 of type dbl 5% 0 0 0 0 0 0 0 0 0 0 ⋯ 0 0 0 0 0 0.000 0 0 0 0 94% 11 4 1 1 2 2 0 0 1 3 ⋯ 30 1 4 1 7 10.055 0 3 1 1
12H7. In the trolley data — data(Trolley) — we saw how education level (modeled as an ordered
category) is associated with responses. But is this association causal? One plausible confound is
that education is also associated with age, through a causal process: People are older when they
finish school than when they begin it. Reconsider the Trolley data in this light. Draw a DAG that
represents hypothetical causal relationships among response, education, and age. Which statical
model or models do you need to evaluate the causal influence of education on responses? Fit these
models to the trolley data. What do you conclude about the causal relationships among these three
variables?
ERROR:
Which statical [sic] model or models do you need
Answer. Consider the following DAGs. Implicit in this list of DAGs is that the response would
never influence age or education. Because we already know that education is associated with the
response, we also only include DAGs where Education and Response are d-connected.
A more difficult question is what arrows to allow between education and age. Getting older does not
necessarily lead to more education, but education always leads to greater age. That is, someone
could stop schooling at any point in their lives. This perspective implies Education should cause
Age, because the choice to get a particular education is also a choice to get older before
continuing with ‘real’ work.
The issue with allowing this arrow is it contradicts the following definition of Causal dependence from David Lewis:
An event E causally depends on C if, and only if, (i) if C had occurred, then E would have occurred, and (ii) if C had not occurred, then E would not have occurred.
Said another way, causality is almost always associated with the advancement of time. This is
inherent in the definition of causality above, and in lists (see #4) such as the Bradford-Hill
criteria. See also the definition of Causal inference. The Age covariate is confusingly
a measurement of time. It is perhaps best to think of Age as a measurement of the potential to get
education; greater Age indicates an individual is ‘richer’ in terms of time opportunity to get an
education.
Said yet another way, age is not controllable. We will get older regardless of whether we get an education. Education on the other hand is clearly within our control. The issue seems to be that we require two things to get a particular educational degree: a personal commitment to do so and an investment of our time. We talk about investing our time when in the end we are always forced to invest it in something; we can’t choose to stay in a frozen present.
Said yet another way, we should not confuse prediction and causal inference. Someone’s education may help us predict their age, but this does not imply the relationship is causal.
Described as a rule for DAGs, indirect measures of time (like Age) can only have arrows pointing out
of them. A convenient side effect of this rule is that to get the total causal effect of a variable
like Age there should never be any back door paths to close.
Do these rules contradict general relativity? If there was a variable for how fast or far someone had traveled in their life (e.g. if they were an astronaut) couldn’t that cause a change in their Age? Perhaps in this extra special case you’d need to define a standardized time that all other time measurements were made relative to.
This perspective leads to other questions. Age does not necessarily indicate the level of education we will eventually achieve. Are people who had a healthy upbringing, e.g. with more material resources, more or less likely to provide disapproving responses, and also more or less likely to get an education? There are plenty of possible confounds that could influence both eventual education level and response, such as temperament. Would a 14 year old who will eventually get a PhD answer the same way to these questions now and after they finish their PhD?
Practially speaking, based on the wording in the question and the last paragraph of section 12.4, it
seems the author wanted readers to only consider that Age influences Education (not vice versa).
See also the next question, which adds a few more comments on this issue.
Conditional independencies, if any, are listed under each DAG.
In this first model, education is a mediator between age and response:
library(dagitty)
age_dag <- dagitty('
dag {
bb="0,0,1,1"
Age [pos="0.4,0.2"]
Education [pos="0.6,0.2"]
Response [pos="0.5,0.3"]
Age -> Education
Education -> Response
}')
iplot(function() plot(age_dag), scale=10)
display(impliedConditionalIndependencies(age_dag))
Age _||_ Rspn | Edct
In this model, all variables are associated regardless of conditioning:
mediator_dag <- dagitty('
dag {
bb="0,0,1,1"
Age [pos="0.4,0.2"]
Education [pos="0.6,0.2"]
Response [pos="0.5,0.3"]
Age -> Education
Age -> Response
Education -> Response
}')
iplot(function() plot(mediator_dag), scale=10)
display(impliedConditionalIndependencies(mediator_dag))
In this model, education has no direct causal effect on the response:
independent_dag <- dagitty('
dag {
bb="0,0,1,1"
Age [pos="0.4,0.2"]
Education [pos="0.6,0.2"]
Response [pos="0.5,0.3"]
Age -> Education
Age -> Response
}')
iplot(function() plot(independent_dag), scale=10)
display(impliedConditionalIndependencies(independent_dag))
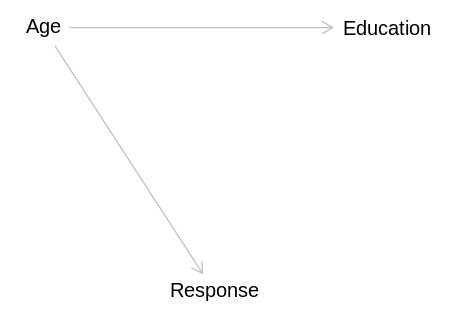
Edct _||_ Rspn | Age
This answer will not consider this model because, as stated above, it seems more reasonable to
assume that age leads to education than vice versa. Notice it has the same conditional
independencies as the last model, and we could construct similar models where Education -> Age and
with the same conditional independencies as the first and second models. Although that approach
would work for this question, see comments in the next question.
independent_dag <- dagitty('
dag {
bb="0,0,1,1"
Age [pos="0.4,0.2"]
Education [pos="0.6,0.2"]
Response [pos="0.5,0.3"]
Education -> Age
Age -> Response
}')
iplot(function() plot(independent_dag), scale=10)
display(impliedConditionalIndependencies(independent_dag))
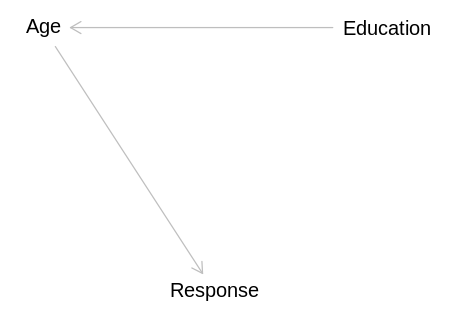
Edct _||_ Rspn | Age
To test which model is correct, we only need one more model beyond what we learned from m12.6 in
the text: a model that includes age as a predictor. If we find that the education coefficient bE
is near zero in this model, the third causal model is correct. If we find that the age coefficient
is near zero, the first causal model is correct. If both coefficients are non-zero, we can infer the
second causal model is correct. Of course, all causal inferences are limited by the variables we are
considering.
data(Trolley)
d <- Trolley
edu_levels <- c(6, 1, 8, 4, 7, 2, 5, 3)
d$edu_new <- edu_levels[d$edu]
dat <- list(
R = d$response,
action = d$action,
intention = d$intention,
contact = d$contact,
E = as.integer(d$edu_new),
StdAge = standardize(d$age),
alpha = rep(2, 7)
)
m_trolley_age <- ulam(
alist(
R ~ ordered_logistic(phi, kappa),
phi <- bE * sum(delta_j[1:E]) + bStdAge * StdAge + bA * action + bI * intention + bC * contact,
kappa ~ normal(0, 1.5),
c(bA, bI, bC, bE, bStdAge) ~ normal(0, 1),
vector[8]:delta_j <<- append_row(0, delta),
simplex[7]:delta ~ dirichlet(alpha)
),
data = dat, chains = 4, cores = min(detectCores(), 4), iter=2000
)
iplot(function() {
plot(precis(m_trolley_age, depth=2), main="m_trolley_age")
}, ar=1.8)
display_markdown("Raw data (preceding plot):")
display(precis(m_trolley_age, depth = 2), mimetypes="text/plain")
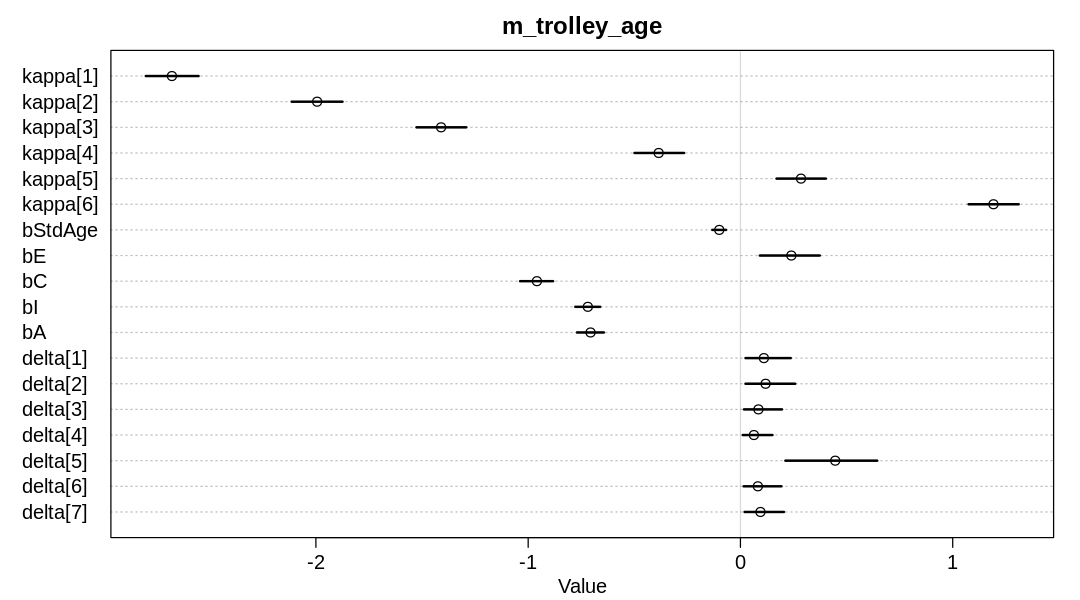
Raw data (preceding plot):
mean sd 5.5% 94.5% n_eff Rhat4
kappa[1] -2.67830561 0.08343011 -2.80122645 -2.55278302 1078.6704 1.0024045
kappa[2] -1.99419504 0.08108227 -2.11397697 -1.87468153 1028.9019 1.0025716
kappa[3] -1.41015518 0.07981220 -1.52607675 -1.29153444 1003.5752 1.0027147
kappa[4] -0.38516554 0.07943401 -0.49912643 -0.26571646 1009.6277 1.0031024
kappa[5] 0.28527392 0.07997250 0.17009254 0.40234439 988.9081 1.0032235
kappa[6] 1.19187995 0.08146554 1.07484372 1.31077185 1030.7310 1.0029348
bStdAge -0.09980456 0.02057290 -0.13243610 -0.06778931 2478.8851 1.0011160
bE 0.23973082 0.09967973 0.09144668 0.37359848 824.1898 1.0039400
bC -0.95887192 0.04860664 -1.03782058 -0.88270538 3987.0337 0.9993633
bI -0.71895204 0.03683421 -0.77789181 -0.66061573 4279.5796 1.0002961
bA -0.70615415 0.04017477 -0.77006368 -0.64310355 3974.8459 0.9997743
delta[1] 0.11041244 0.06987493 0.02386914 0.23702128 4110.5247 1.0002486
delta[2] 0.11840614 0.07534208 0.02369014 0.25798016 5122.1495 0.9994711
delta[3] 0.08474526 0.05982338 0.01655209 0.19596407 3165.8438 1.0014157
delta[4] 0.06340311 0.04964455 0.01127020 0.15086570 1929.8542 1.0011308
delta[5] 0.44650719 0.13548407 0.21163334 0.64445624 1373.2541 1.0020109
delta[6] 0.08217264 0.05801588 0.01511948 0.19316777 4390.0564 0.9994020
delta[7] 0.09435321 0.06108534 0.01951332 0.20496406 4283.7173 0.9999439
As implied in the text, it is not correct to causally infer the associated of education bE is
negative based on m12.6. In this model, the association of education with response has reversed,
becoming positive. Since the error bars on neither bStdAge or bE cross zero, we’ll infer for now
that the second causal model above is correct.
12H8. Consider one more variable in the trolley data: Gender. Suppose that gender might influence education as well as response directly. Draw the DAG now that includes response, education, age, and gender. Using only the DAG, is it possible that the inferences from 12H7 above are confounded by gender? If so, define any additional models you need to infer the causal influence of education on response. What do you conclude?
Answer. First, lets consider a DAG that doesn’t work. In the following DAG we assume
Education -> Age rather than that Age -> Gender:
incorrect_dag <- dagitty('
dag {
bb="0,0,1,1"
Education [pos="0.4,0.2"]
Age [pos="0.6,0.2"]
Response [pos="0.5,0.3"]
Gender [pos="0.3,0.3"]
Education -> Age
Education -> Response
Age -> Response
Gender -> Education
Gender -> Response
}')
iplot(function() plot(incorrect_dag), scale=10)
display(impliedConditionalIndependencies(incorrect_dag))
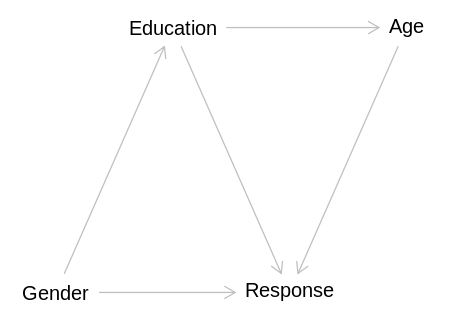
Age _||_ Gndr | Edct
With the additions implied by this question, this DAG implies that Gender will be able to predict
Age without conditioning, which is not correct.
Instead, we’ll add this question’s covariate to the second model from the last question:
gender_dag <- dagitty('
dag {
bb="0,0,1,1"
Age [pos="0.4,0.2"]
Education [pos="0.6,0.2"]
Response [pos="0.5,0.3"]
Gender [pos="0.7,0.3"]
Age -> Education
Education -> Response
Age -> Response
Gender -> Education
Gender -> Response
}')
iplot(function() plot(gender_dag), scale=10)
display(impliedConditionalIndependencies(gender_dag))
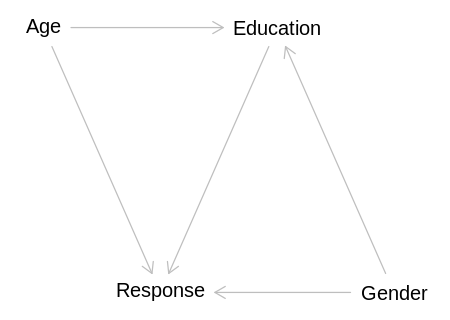
Age _||_ Gndr
This DAG implies Age will always be independent of Gender, which seems reasonable in this
context, ignoring that females live slightly longer than males. To infer either the direct or total
causal effect of Education on Response the required adjustment is to condition on both Age and
Gender:
display(adjustmentSets(gender_dag, exposure="Education", outcome="Response", effect="total"))
{ Age, Gender }
We can do so by fitting a model that includes all three variables as predictors of Response:
data(Trolley)
d <- Trolley
edu_levels <- c(6, 1, 8, 4, 7, 2, 5, 3)
d$edu_new <- edu_levels[d$edu]
dat <- list(
R = d$response,
action = d$action,
intention = d$intention,
contact = d$contact,
E = as.integer(d$edu_new),
StdAge = standardize(d$age),
gid = ifelse(d$male == 1, 2, 1),
alpha = rep(2, 7)
)
# This fit takes more than three hours with 4 chains/cores.
m_trolley_gender <- ulam(
alist(
R ~ ordered_logistic(phi, kappa),
phi <- bE * sum(delta_j[1:E]) + bStdAge * StdAge + bGender[gid]
+ bA * action + bI * intention + bC * contact,
kappa ~ normal(0, 1.5),
bGender[gid] ~ normal(0, 1),
c(bA, bI, bC, bE, bStdAge) ~ normal(0, 1),
vector[8]:delta_j <<- append_row(0, delta),
simplex[7]:delta ~ dirichlet(alpha)
),
data = dat, chains = 2, cores = min(detectCores(), 2), iter=1600
)
flush.console()
iplot(function() {
plot(precis(m_trolley_gender, depth=2), main="m_trolley_gender")
}, ar=1.8)
display_markdown("Raw data (preceding plot):")
display(precis(m_trolley_gender, depth = 2), mimetypes="text/plain")
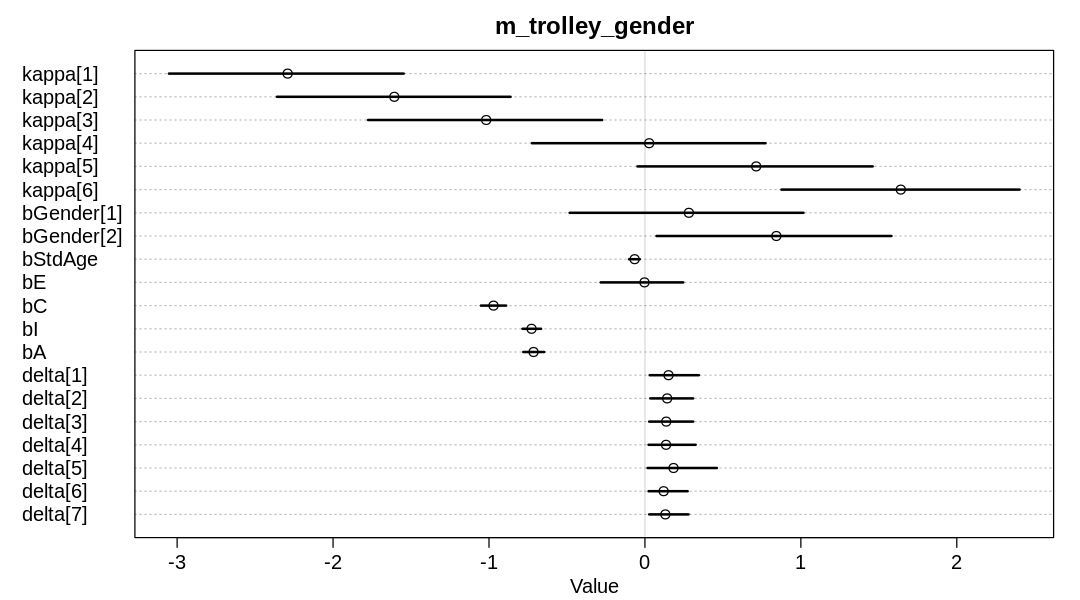
Raw data (preceding plot):
mean sd 5.5% 94.5% n_eff Rhat4
kappa[1] -2.291288958 0.46714983 -3.05260629 -1.54696264 659.4706 0.9999423
kappa[2] -1.607263559 0.46604267 -2.36025842 -0.86161796 651.3348 0.9999119
kappa[3] -1.017986406 0.46601190 -1.77572631 -0.27447415 646.3224 0.9999655
kappa[4] 0.027295827 0.46724972 -0.72466027 0.77361815 650.8136 0.9999874
kappa[5] 0.713412755 0.46689327 -0.04813372 1.46074243 646.8730 1.0000190
kappa[6] 1.640855013 0.46595949 0.87522556 2.40258796 656.7113 1.0000113
bGender[1] 0.281718757 0.47214155 -0.48282535 1.01670089 648.9819 1.0008279
bGender[2] 0.842675884 0.47292313 0.07431572 1.58122866 650.7906 1.0007947
bStdAge -0.065964224 0.02168272 -0.10192004 -0.03280363 656.3598 1.0031312
bE -0.002843373 0.17313847 -0.28393684 0.24594105 432.4367 1.0069854
bC -0.971089462 0.05088699 -1.05192433 -0.89012013 1163.9056 1.0005626
bI -0.727087854 0.03724670 -0.78542080 -0.66563698 1484.6136 1.0001663
bA -0.714202313 0.04216100 -0.78006046 -0.64593394 1198.0312 1.0014116
delta[1] 0.151104519 0.10052852 0.03023021 0.34580260 1200.1489 0.9999488
delta[2] 0.142577365 0.08988162 0.03408959 0.30904329 1532.9985 0.9996467
delta[3] 0.136646640 0.09215948 0.02706038 0.30950045 1307.6900 1.0005605
delta[4] 0.135418403 0.09860457 0.02332306 0.32535805 891.6557 1.0019536
delta[5] 0.183354260 0.15339020 0.01686351 0.46168231 533.0076 1.0071922
delta[6] 0.119878490 0.07915463 0.02342631 0.27391009 1886.3322 0.9990988
delta[7] 0.131020323 0.08137387 0.02700134 0.27988752 1990.5208 1.0010292
At this point, education appears to have no causal effect on response. That is, the mean and HPDI of
bE are near zero.